Molecular insights to crustacean phylogeny
The PhD project. My PhD project was integrated in the priority program “Deep metazoan Phylogeny” (http://www.deep-phylogeny.org) of the DFG (German Science Foundation).
Goals. I aimed to 1) contribute to the reconstruction of the phylogenetic relationships within the crustaceans that are still very unclear and 2) to reaveal the postition of the crustaceans within the arthropods. Important for these aims is a good taxon sampling, consequently a large part of my PhD thesis comprised fieldwork and the collection of special crustacean groups, like the Remipedia, Mystacocarida, Pentastomida etc. This demanded scuba diving and research vessel based collection trips.
General methods. I used 1) standard genes [12S rRNA, 16S rRNA, 18S rRNA, 28S rRNA, COI, Histon 3] and 2) later large scale and next generation sequences (ESTs) to reconstruct molecular phylogenies. A major focus and aspect in the analyses was the alignment procedure and the data quality, addressing also the impact of complex modelling on the data analyses.
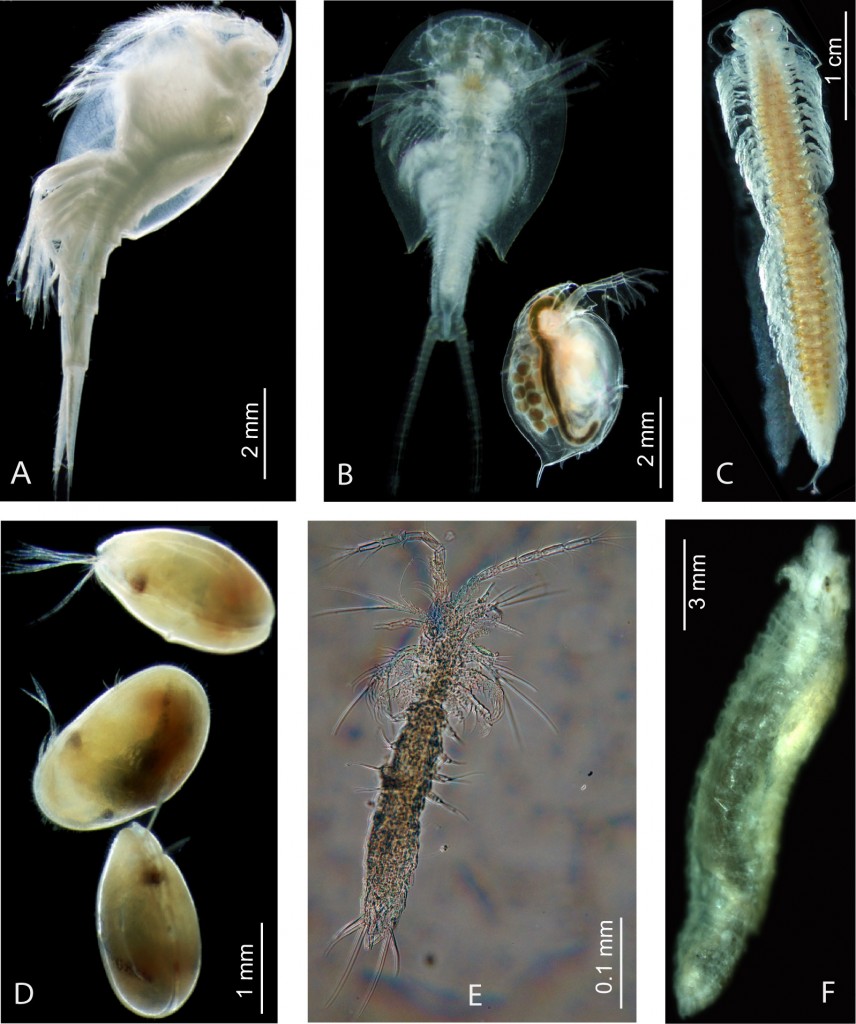
Conclusions. House keeping gene and multi gene analyses are obviously not capable to reveal crustacean or pancrustacean phylogeny, and to an extent also arthropod phylogeny. The study of Koenemann et al. (2010) shows clearly how limited the phylogenetic analyses with the standard house keeping genes are. In the methodological study of rRNA genes it could be demonstrated empirically, that inhomogeneous base compositions and heterogeneous substitution processes influence phylogenetic inference. However, one positive outcome was that it was shown that Cephalocarida are dramatically mis-positioned in analyses that do not implement methods to prevent these effects. A clade Cephalocarida + Remipedia was yet doubted by Spears and Abele, when they discussed this grouping in their reconstructed trees. The graphics (above) show some of the crustacean diversity: A) Leptostraca, B) Branchiopoda, C) Remipedia, D) OStracoda, E) Mystacocarida, F) Pentastomida.
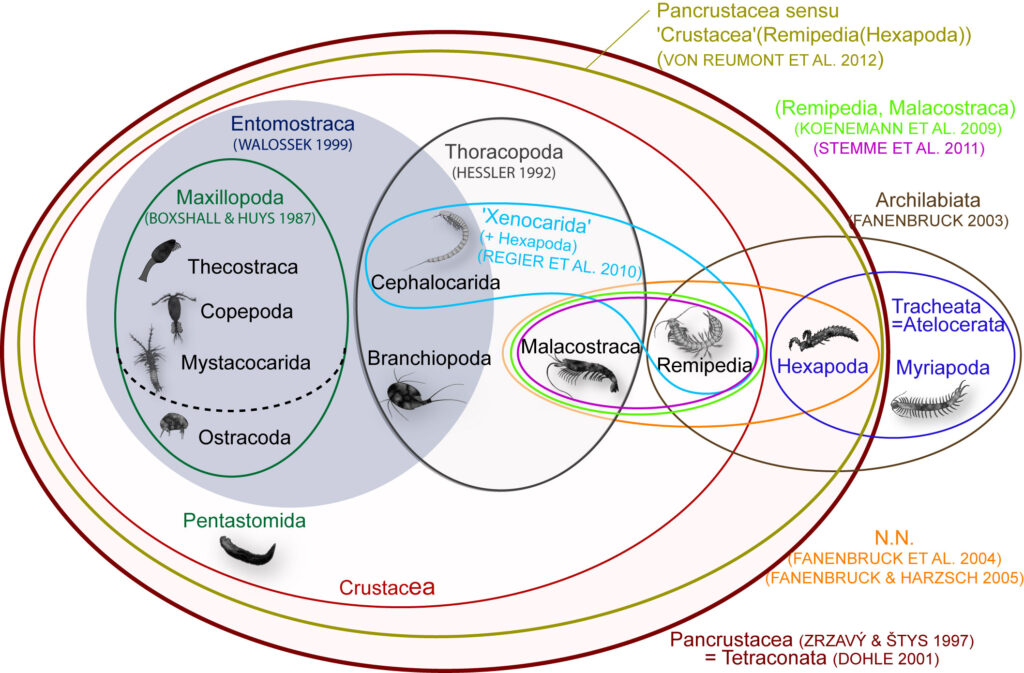 Conflicting morphological hypotheses. The Venn diagram (right) shows the ambiguous phylogeny hypotheses that address crustacean evolution based on morphological data. The incredible diversity and variation in body shapes is definitively one reason for difficulties to reconstruct crustacean phylogeny based on morphological characters.
Conflicting morphological hypotheses. The Venn diagram (right) shows the ambiguous phylogeny hypotheses that address crustacean evolution based on morphological data. The incredible diversity and variation in body shapes is definitively one reason for difficulties to reconstruct crustacean phylogeny based on morphological characters.
Perspective. The hope is, that transcriptomic data in contrast to single or multi gene studies might be capable to resolve crustacean phylogeny. However, crustacean phylogenomics is in many aspects still in its beginnings, starting with a still incomplete taxon sampling. See also current projects, pancrustacean phylogenomics. Still open questions on internal crustacean phylogeny are only to tackle and to address in framework of a larger consortium similar to the 1KITE initiative that unites experts on all crustacean taxa and of molecular and morphological expertise.