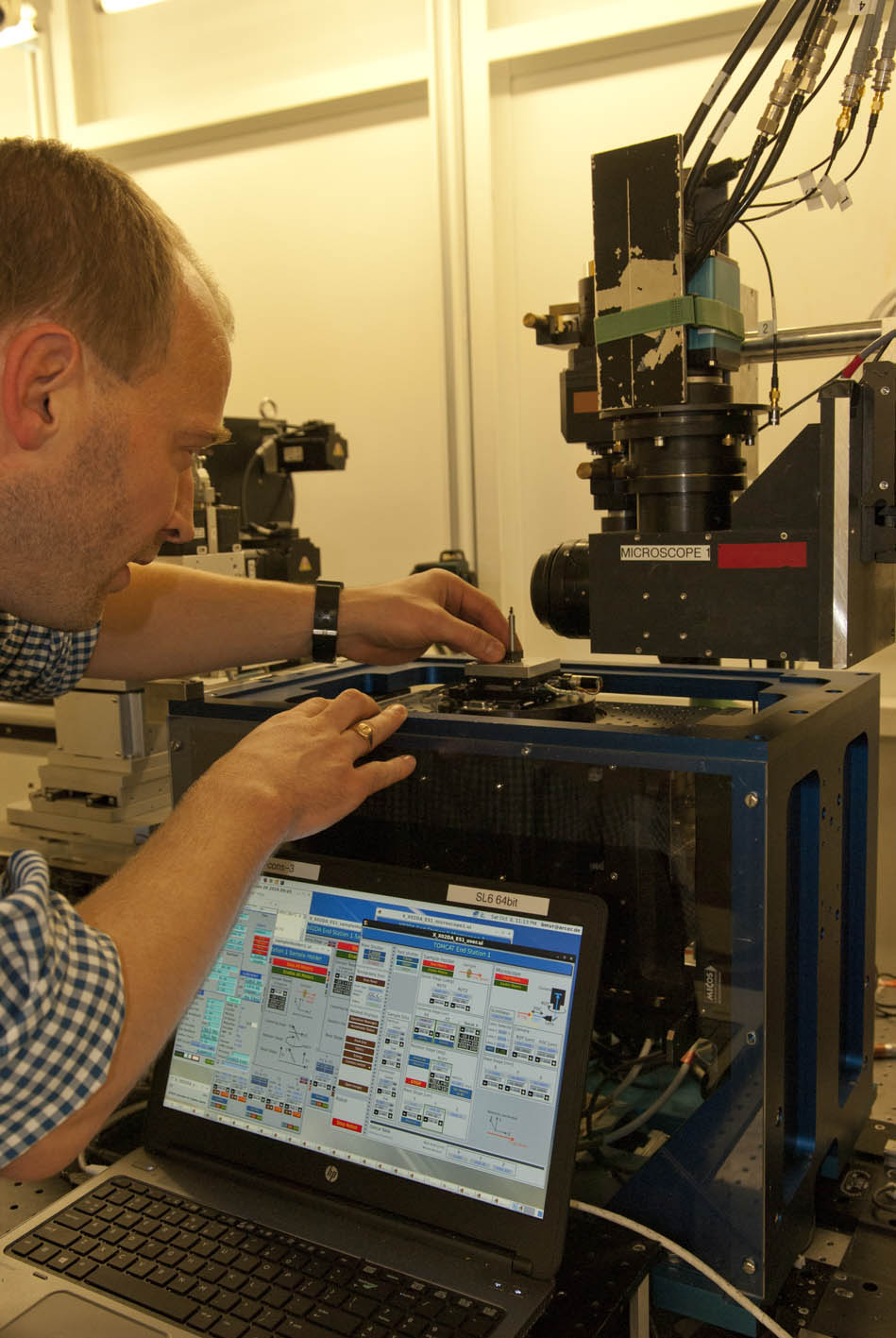
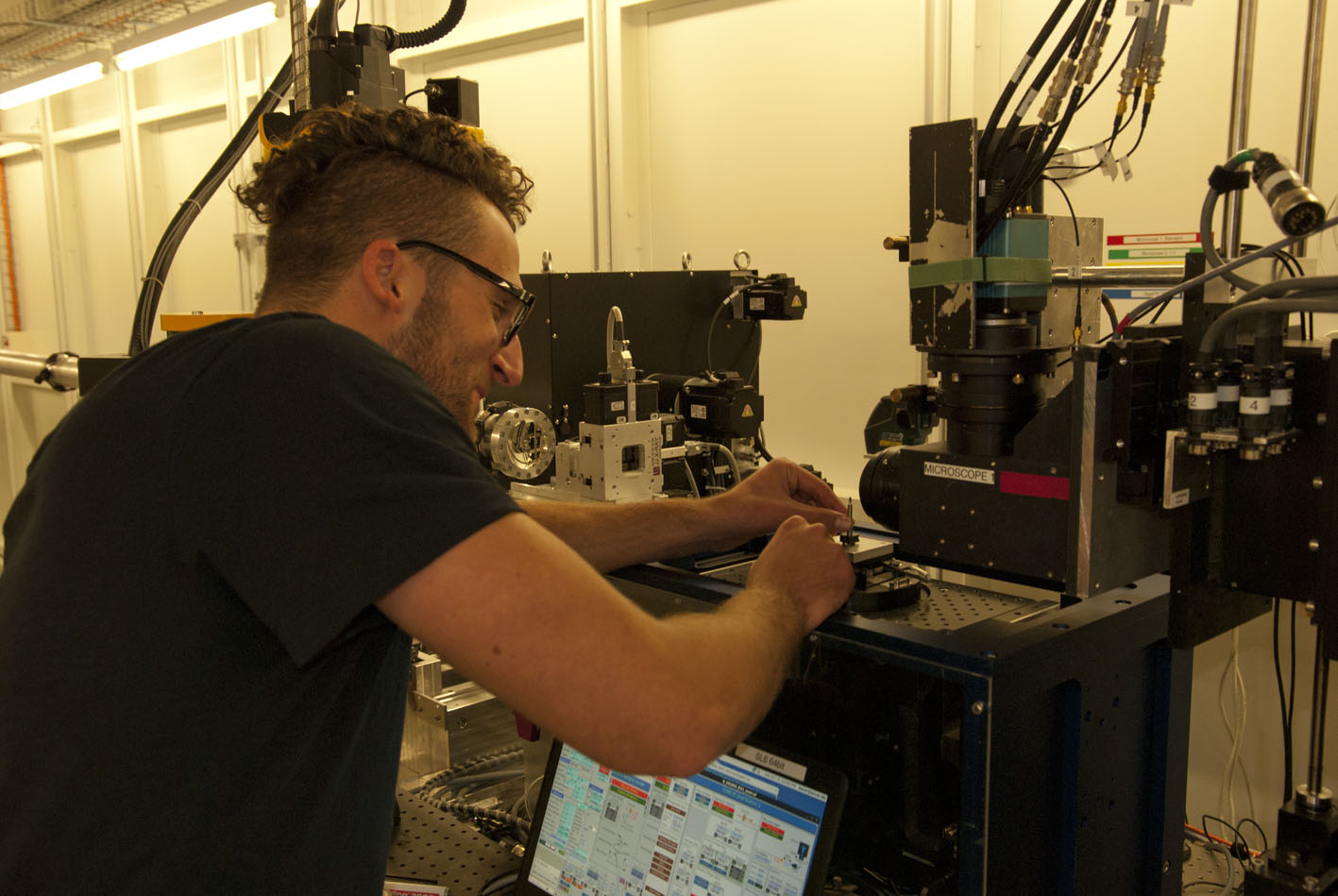
Species collection
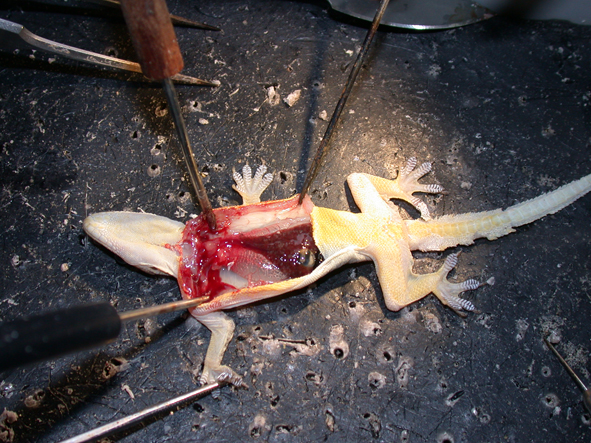
I collect as many specimens by myself. This includes sampling of marine organisms (benthic, pelagic and meiofauna using Van Veen grabber, multicore, dredging, research vessels, scuba diving) but also terrestrial species (diverse netting methods, berlese- malaise- pitfall-traps). My motivation to work in evolutionary biology is also driven by the curiosity in diversity and biology of species and to understand their biology and ecology better. I try to awaken this fascination also in my students, which starts with species collection.
Laboratory work
I worked extensively in dry and wet labs. Classic DNA based molecular lab work started with primer design, PCR amplification and sequencing of gene fragments for single and multigene analyses. I worked on old LI COR and ABI 377 plate sequencers and established and used later on sequencing protocols on different Beckman Coulter capillary sequencers. Starting with EST and later next generation sequencing data, my laboratory skills were extended to RNA based work using different types of RNA extraction (Trizol and kit based) methods and cDNA library amplification and sequencing.
Phylogenetics / Phylogenomics
My background and original training is in molecular evolutionary biology, and thus I was and am involved in the application and constant improvement of molecular based methods adapting to new technology. This includes single, multigene and transcriptome based (phylogenomic) analyses. In phylogenomics and genomics I have strong collaborations with consortia such as 1KITE and i5K to tackle this challenge and to push boundaries for large scale data analyses.
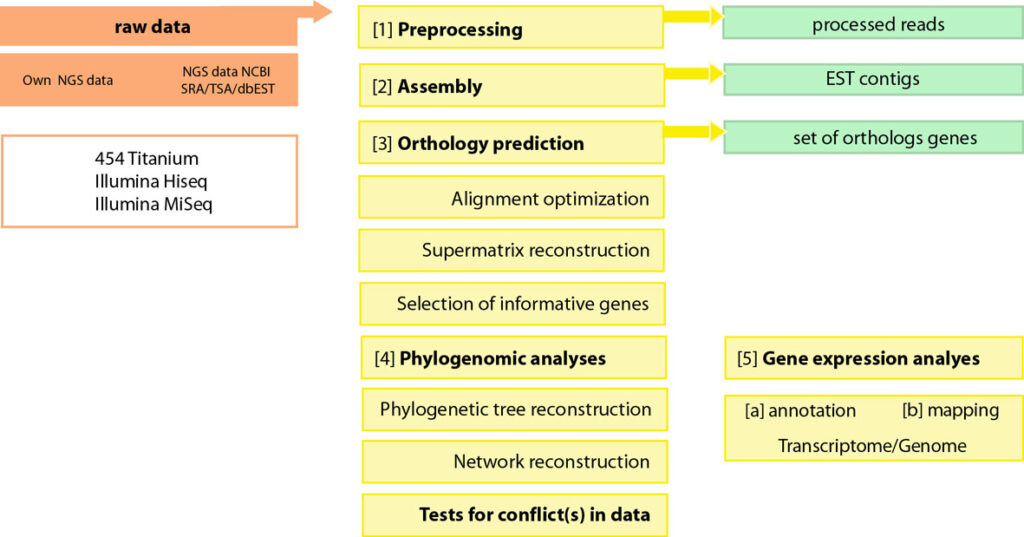
Functional morphology
The evolution and anatomy of venom delivery systems for neglected or obsure venomous taxa is often unclear, particular for invertebrate species. To understand the anatomy of venom delivery systems better and to enlight their possible functional morphology I apply state of the art synchrotron based miro-computer tomography 3D reconstructions. Of course open source software is used for that.
Transcriptomics (in venomics)
I utilize transcriptomics for classic phylogenomics but recently in particular for venomics. To understand venom evolution transcriptome data is used to analyze gene expression levels in combination with protomic data. In most cases for small invertebrates the workflow (von Reumont etal. 2017) differs slightly, and a differential gene expression applying replicates of venom gland transcriptome samples is not possible.
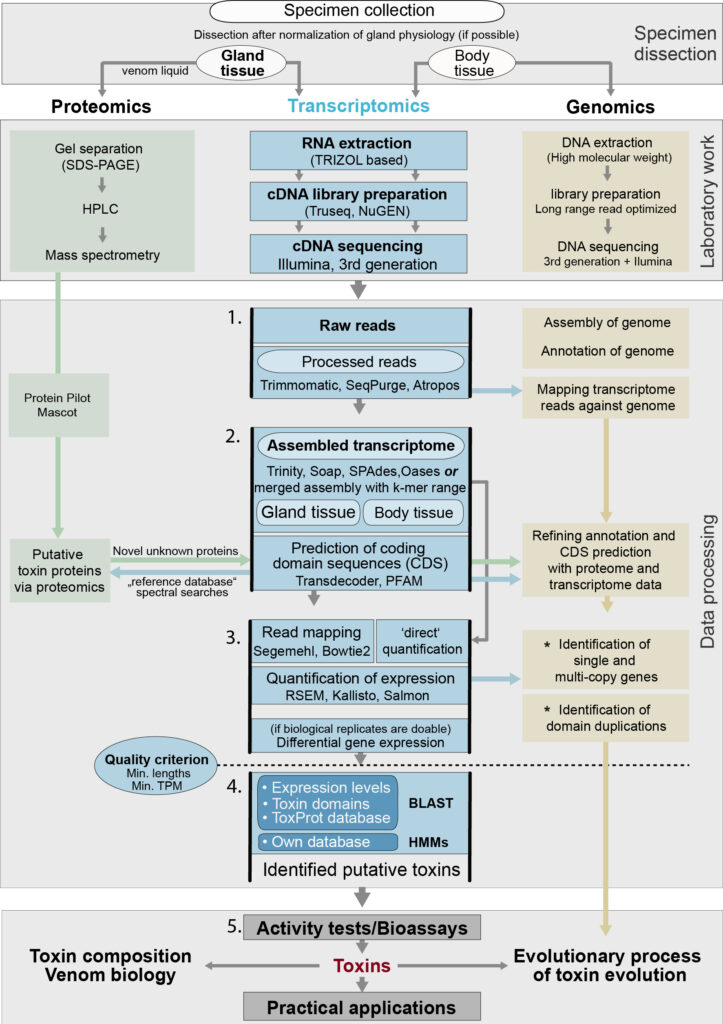
Comparative genomics (integrating proteomics, transcriptomics)
I am interested in genomics from a twofold perspective. Within a phylogenomic context I like to understand general evolution of pancrustaceans for example the transition from marine to terrestrial habitats. Linked to venomics genome data of venomous species is important to understand the processes that drive evolution of toxins. I have strong collaborations within the i5K initiative, the ZMB at the Researchmuseum Alexander Koenig Bonn and in LOEWE TBG (LOEWE Center for Translational Biodiversitygenomics) to work on genomics of hymenopteran and pancrustacean species. Several species are currently being sequenced using the technology and platforms of PacBio, Nanopore and Illumina platforms. Transcriptome and proteome data are combined with genomics to predict proteins and genes. Currently I have strong collaborations to analyze proteome data with experts in Germany (Gnther Lochnit, JLU), Australia (Eivind Undheim) and France (Sebastien Dutertre).
Artificial Intelligence – Machine Learning (Neural networks)
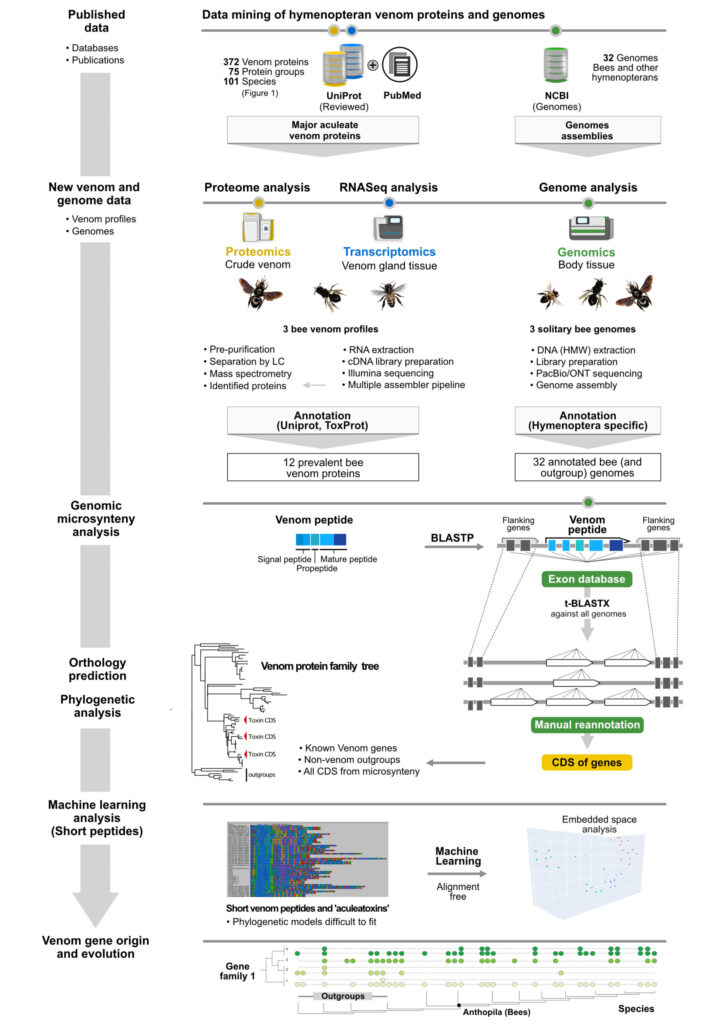
Being recently trained as a data scientist, I utilize deep learning to analyse proteins and genes. Large language models do the trick here to conclude on phylogenetic and functional relation. The shown workflow illustrates a recent analysis on bee venom genes using multi-omics methods combined with a novel machine learning approach.