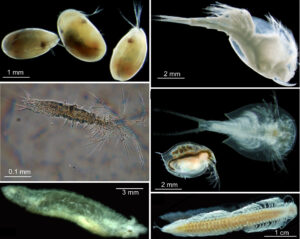
| • Molecular phylogeny of crustaceans and eu-arthropods based on multiple gene analyses I started to work on crustacean, pancrustacean and euarthropod phylogeny in my PhD thesis. Basically starting from scratch I collected all non-malacostracan species by myself, like Pentastomida, Mystacocarida, Remipedia etc. However, it was kind of frustrating or enlighting (depending on your own philosophy, I am more the positive person and prefer latter perspective) that even with most complex modeling respectively the crustacean phylogeny was not clearly to recover with multi gene analyses despite that intensive, complete taxon sampling. |
 |
| • Phylogeography of the species complex Zygaena angelica/transalpina Speciation is one of the topics that fascinates me. I was lucky to start working in this field with my master thesis and could finally extent and finish this study on diurnal zygaenid moths (Zygaenida, Lepidoptera) during my PhD. The species complex is, as many central european species, influenced by the effects during the last glacial. We could show that complex separation patterns and micro-refugia played obviously an important role to drive speciation. |
|
| • Diversity and monitoring of the local bat fauna of Bonn and the limestone quarries “Ofenkaulen”, Germany My first contact to cave systems was terrestrial and reasoned by monitoring overwintering bats in local quarry remainings and mines in the mountain range “Siebengebirge” near Bonn. Additionally, I organized and planned as one of the founding members of the local bat research group (BAFF) monitoring by netting and detector work to describe and monitor the diversity of the local bat fauna. |

|
| • Primer-Tool-Box development for arthropods based on EST data During my PhD degenerated primer sets were developed to sequence informative genes for arthropod subgroups based on EST data and known genes. This approach was extensively tested for crustaceans, however one outcome was that crustaceans are so diverse that primers sets had to be adapted manually in subsequent PCRs, which is extremely time-intensive. The project was collaboratively conducted with theworking groups of Bernahrd Misof (Bonn, Germany), Günther Pass and Elisabeth Haring (Vienna, Austria). |
